It is now easier to pinpoint exactly what molecules a phosphatase – a type of protein that's essential for cells to react to their environment – acts upon in human cells, thanks to the free online database DEPOD, created by EMBL scientists. Published today in Science Signaling, the overview of interactions could even help explain unforeseen side-effects of drugs.
Although we know the tool's general purpose, it can sometimes be difficult to tell if a specific pair of precision tweezers belongs to a surgeon or a master jeweller. It is now easier to solve similar conundrums about a type of protein that allows cells to react to their environment, thanks to scientists at the European Molecular Biology Laboratory (EMBL). Published today in Science Signaling, their work offers a valuable resource for other researchers.
Whether in your eye being hit by light, in your blood fighting off disease, or elsewhere throughout your body, cells have to react to changes in their environment. But first, a cell must 'know' the environment has changed. One of the ways in which that information is transmitted within the cell is through tags called phosphate ions, which are added to or removed from specific molecules depending on the exact message that has to be conveyed. The tools the cell uses to remove phosphate ions are proteins called phosphatases. But it's not always obvious what molecules – or substrates – a particular phosphatase acts upon.
ESIMERKKI
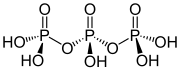
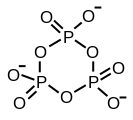
Ajattele inositolifosfaattien ja fosfoinositidien varaan rakennettua kehomme signaalijärjestelmää kuin hienona kellon koneistona! Jos sellaiseen mättää inositolijärjestelmälle turtaa muuta fosfaattia- tarkka metbolinen ja signaloiva ajastin tukkeutuu
--

"One of the biggest
challenges in phosphatase research is finding substrates, and this is
what our work supports," says Maja Köhn from EMBL in Heidelberg,
Germany, who led the study. "We've made it easier to create hypotheses
about the relationships between phosphatases and their substrates."
Xun Li, a post-doctoral student shared by Köhn's lab and those of Matthias Wilmanns at EMBL in Hamburg, Germany and Janet Thornton at EMBL-European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK, compiled the most complete picture to date of all the phosphatases in human cells, and their substrates. The scientists also grouped phosphatases into families, based on their three-dimensional structure, which can influence what molecules a phosphatase can act upon.
This information allows researchers to easily identify a phosphatase's known substrates, and suggest new substrates based on how similar it is to other phosphatases. The web-like overview of interactions could even help explain unforeseen side-effects of drugs designed to interfere with phosphatases or with their phosphate-adding counterparts, kinases. To enable others to make such connections, Köhn and colleagues have created a free online database, DEPOD.
"When people have unexpected results, this could be a place to find explanations," says Thornton, head of EMBL-EBI. "DEPOD combines a wealth of information that can be explored in a variety of ways, to make it useful not just to phosphatase researchers but to the wider community."
Read more at: http://phys.org/news/2013-05-database-phosphate-substrate.html#jCp
Xun Li, a post-doctoral student shared by Köhn's lab and those of Matthias Wilmanns at EMBL in Hamburg, Germany and Janet Thornton at EMBL-European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK, compiled the most complete picture to date of all the phosphatases in human cells, and their substrates. The scientists also grouped phosphatases into families, based on their three-dimensional structure, which can influence what molecules a phosphatase can act upon.
This information allows researchers to easily identify a phosphatase's known substrates, and suggest new substrates based on how similar it is to other phosphatases. The web-like overview of interactions could even help explain unforeseen side-effects of drugs designed to interfere with phosphatases or with their phosphate-adding counterparts, kinases. To enable others to make such connections, Köhn and colleagues have created a free online database, DEPOD.
"When people have unexpected results, this could be a place to find explanations," says Thornton, head of EMBL-EBI. "DEPOD combines a wealth of information that can be explored in a variety of ways, to make it useful not just to phosphatase researchers but to the wider community."
Read more at: http://phys.org/news/2013-05-database-phosphate-substrate.html#jCp
"One of the biggest
challenges in phosphatase research is finding substrates, and this is
what our work supports," says Maja Köhn from EMBL in Heidelberg,
Germany, who led the study. "We've made it easier to create hypotheses
about the relationships between phosphatases and their substrates."
Xun Li, a post-doctoral student shared by Köhn's lab and those of Matthias Wilmanns at EMBL in Hamburg, Germany and Janet Thornton at EMBL-European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK, compiled the most complete picture to date of all the phosphatases in human cells, and their substrates. The scientists also grouped phosphatases into families, based on their three-dimensional structure, which can influence what molecules a phosphatase can act upon.
This information allows researchers to easily identify a phosphatase's known substrates, and suggest new substrates based on how similar it is to other phosphatases. The web-like overview of interactions could even help explain unforeseen side-effects of drugs designed to interfere with phosphatases or with their phosphate-adding counterparts, kinases. To enable others to make such connections, Köhn and colleagues have created a free online database, DEPOD.
"When people have unexpected results, this could be a place to find explanations," says Thornton, head of EMBL-EBI. "DEPOD combines a wealth of information that can be explored in a variety of ways, to make it useful not just to phosphatase researchers but to the wider community."
Read more at: http://phys.org/news/2013-05-database-phosphate-substrate.html#jCp
Xun Li, a post-doctoral student shared by Köhn's lab and those of Matthias Wilmanns at EMBL in Hamburg, Germany and Janet Thornton at EMBL-European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK, compiled the most complete picture to date of all the phosphatases in human cells, and their substrates. The scientists also grouped phosphatases into families, based on their three-dimensional structure, which can influence what molecules a phosphatase can act upon.
This information allows researchers to easily identify a phosphatase's known substrates, and suggest new substrates based on how similar it is to other phosphatases. The web-like overview of interactions could even help explain unforeseen side-effects of drugs designed to interfere with phosphatases or with their phosphate-adding counterparts, kinases. To enable others to make such connections, Köhn and colleagues have created a free online database, DEPOD.
"When people have unexpected results, this could be a place to find explanations," says Thornton, head of EMBL-EBI. "DEPOD combines a wealth of information that can be explored in a variety of ways, to make it useful not just to phosphatase researchers but to the wider community."
Read more at: http://phys.org/news/2013-05-database-phosphate-substrate.html#jCp



 6.6
6.6





